Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
Create Selection with Mouse in PyMol
This section provides a tutorial on how to create a new selection by selecting atoms with mouse in a molecule in PyMol.
You can use PyMol to visualize a molecule structure as a whole. Or you can define sub-sets of the molecule as selections and manage visualization of each selection independently.
The first step to create a selection is to select atoms from the molecule to be included in the selection.
The simplest way to select atoms is click their locations on the screen and name the selection.
1. Load the molecule from the SDF file, Molecule-HY-001.sdf. You see the molecule in the default representation.
2. Change the representation to "lines" with two commands:
hide all show lines
3. Click on "Selecting" mode to change it to "Atoms". The "Selecting" mode is located in the mouse control area near the bottom right corner of the viewer window.
4. Click on all 6 C atoms on the benzene ring in the lower part of the molecule. You see selected atoms identified with small pink squares. You also see a new entry called "(sele)" displayed on the right side of the viewer window.
5. Click "A" on the "(sele)" entry to open the action menu. And click "rename selection" from the menu. You see a rename message on the viewer window.
6. Enter "ring" as the name of the object created with those selected atoms. You can also rename the "sele" to "ring" with the "set_name sele, ring" command.
Now we have create a new selection called "ring" representing a sub-set of the molecule.
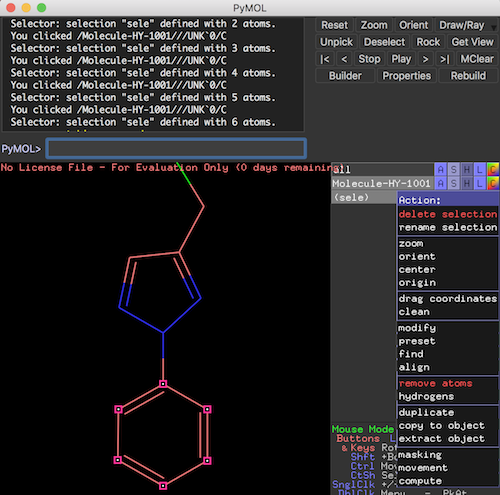
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
►Create Selection with Mouse in PyMol
Substructure Selection Visualization in PyMol
Modify Molecule Structure in PyMol
Export Molecule Substructure in PyMol
Create Methane Molecule in PyMol
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)