Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
PyMol Screen Layout
This section provides a quick introduction of the PyMol screen layout.
Once PyMol is started, you should see two windows displayed:
External GUI - A Graphical User Interface window with the following areas:
1. Output message area - An area where output messages are displayed. Initially, the output area displays the following message:
Copyright (C) Schrodinger, LLC This Executable Build integrates and extends Open-Source PyMOL. Detected OpenGL version 2.1. Shaders available. Detected GLSL version 1.20. OpenGL graphics engine: GL_VENDOR: ATI Technologies Inc. GL_RENDERER: AMD Radeon Pro 450 OpenGL Engine GL_VERSION: 2.1 ATI-1.51.8 No License File - For Evaluation Only (23 days remaining) Detected 8 CPU cores. Enabled multithreaded rendering.
2. Command input area - An area below the output message area where PyMol commands can be entered.
3. Command button area - An area on the right side of the output message area where a set of command buttons are displayed.
Viewer Window - A graphical display window with the following areas:
1. Viewer area - An area where the molecule is rendered as a graphical image. Initially, the molecule viewer displays a blank area.
2. Command input area - An area below the viewer area where PyMol commands can be entered.
3. Object name area - An area where a list of objects identified in the molecule is displayed. Initially, the object area is empty.
4. Mouse information area - An area where help information are displayed to tell you how to control the visualization with the mouse.
5. Movie control area - An area where movie editing and playing controls are displayed.
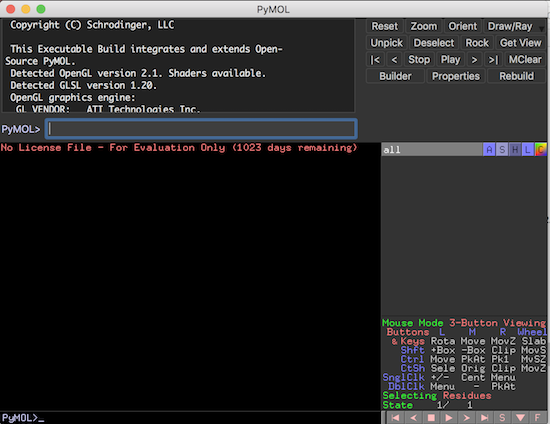
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
Load Molecule from File into PyMol
Virtual Trackball Rotation on PyMol
"load" and "delete" Commands on PyMol
"log_open" and "log_close" Commands on PyMol
Model Space and Camera Space on PyMol
"get_view" and "set_view" on PyMol
View Parameters Auto Adjusted on PyMol
Rotation with Transformation Matrix
Difference of "turn" and "rotate" Commands
Difference of "move" and "translate" Commands
"center", "zoom" and "reset" Commands
Model-to-Camera Space Coordinates Mapping
Camera-to-Model Space Coordinates Mapping
"show lines" Presentation Command
"show sticks" Presentation Command
"show spheres" Presentation Command
"show surface" Presentation Command
"show mesh" Presentation Command
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)