Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
Angle Formed by 3 Atoms in PyMol
This section provides a tutorial on how to measure the angle formed by three picked atoms using the 'get_angle/angle' command in PyMol.
PyMol 4 picked atoms identified as pk1, pk2, pk3 and pk4 selections can also be used to measure the angle formed by 3 atoms.
"get_angle" prints the angle formed by three picked atoms in the message output console:
# print angle formed by 3 atoms identified by pk1, pk2, pk3 get_angle # print angle formed by 3 atoms identified by pk2, pk3, pk4 get_angle pk2, pk3, pk4
"angle" creates a new visualization selection to show the angle formed by three picked atoms:
# create angle (named as ang1) formed by pk1, pk2, pk3 angle ang1 # create angle (named as ang2) formed by pk2, pk3, pk4 angle ang2, pk2, pk3, pk4
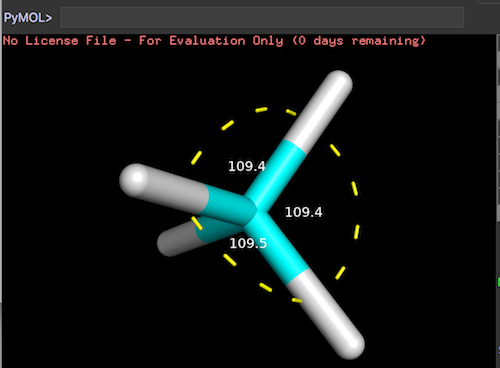
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
"get_extent" - Picked Atom Location
"label" - Generate Labels on Atoms
Distance between Atoms in PyMol
►Angle Formed by 3 Atoms in PyMol
Dihedral Angle Formed by 3 Atoms in PyMol
Use Selection Expressions in PyMol
"get_area" - Surface Areas of Atoms
Surface Area of Entire Molecule
"get_position" - Viewing Center
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)