Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
Model Space and Camera Space on PyMol
This section provides a quick introduction of model space and camera space cooridinate systems used on PyMol to transform and display molecule structures on the screen.
PyMol uses two coordinate systems to form the viewing model when the molecule structure is displayed on the screen.
1. Model Space Coordinate System - A 3-dimensional coordinate system in which molecule atom locations are specified.
For example, the following line in a SDF molecule file specifies a Carbon atom located at (1.1100, 1.2100, 0.0000) in the Model Space coordinate system.
1.1100 1.2100 0.0000 C 0 0 0 0 0 0 0 0
2. Camera Space Coordinate System - A 3-dimensional coordinate system in the display screen and the viewing camera are specified:
- X-axis representing the horizontal direction running from left to right on the screen.
- Y-axis representing the vertical direction running from top to bottom on the screen.
- Z-axis representing the viewing direction running from the viewing camera to the screen.
When you request the molecule structure to be displayed on the screen, PyMol will perform a set of transformation to map the molecule structure from the model space to the camera space. This set of transformations is generated from viewing parameters you specified by viewing commands like "rotate", "move", etc.
The following diagram (source: "A Beginner’s Guide to Molecular Visualization Using PyMOL" By Nicholas Fitzkee) shows you the relation of the model space and the camera space used in PyMol:
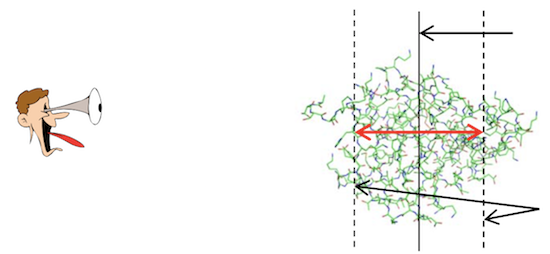
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
Load Molecule from File into PyMol
Virtual Trackball Rotation on PyMol
"load" and "delete" Commands on PyMol
"log_open" and "log_close" Commands on PyMol
►Model Space and Camera Space on PyMol
"get_view" and "set_view" on PyMol
View Parameters Auto Adjusted on PyMol
Rotation with Transformation Matrix
Difference of "turn" and "rotate" Commands
Difference of "move" and "translate" Commands
"center", "zoom" and "reset" Commands
Model-to-Camera Space Coordinates Mapping
Camera-to-Model Space Coordinates Mapping
"show lines" Presentation Command
"show sticks" Presentation Command
"show spheres" Presentation Command
"show surface" Presentation Command
"show mesh" Presentation Command
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)