Molecule Tutorials - Herong's Tutorial Examples - v1.26, by Herong Yang
Modify Molecule Structure in PyMol
This section provides a tutorial on how to modify a molecule structure by rotating and translating a substructure selected from the molecule in PyMol.
Selections in PyMol can be used to visualize substructures differently than the other parts of the molecule. Selections can also be used to modify molecule structure by specifying the selection name in the "rotate" and "translate" commands:
rotate x|y|z degree, selection_name translate [vector], selection_name
Let's continue with the previous tutorial and modify the substructure defined by the "ring" selection.
PyMOL>@Selection-Create-Ring.pml ... PyMOL># show the selection in "sticks" representation PyMOL>show sticks, ring PyMOL># modify substructure "ring" PyMOL>rotate y, 30, ring PyMOL>translate [1,0,0], ring
The molecule structure is modified now as shown on the screen.
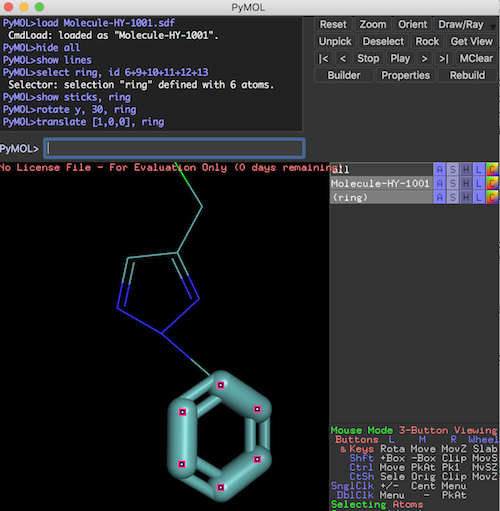
Table of Contents
Molecule Names and Identifications
Nucleobase, Nucleoside, Nucleotide, DNA and RNA
Create Selection with Mouse in PyMol
Substructure Selection Visualization in PyMol
►Modify Molecule Structure in PyMol
Export Molecule Substructure in PyMol
Create Methane Molecule in PyMol
ChEMBL Database - European Molecular Biology Laboratory
PubChem Database - National Library of Medicine
INSDC (International Nucleotide Sequence Database Collaboration)
HGNC (HUGO Gene Nomenclature Committee)